2. Start a Typical Workflow
Qingzhou Zhang
2025-12-03
Source:vignettes/vignette_02-workflow.Rmd
vignette_02-workflow.RmdIntroduction
Protein complexes are the functional machinery of the cell.
High-throughput experimental methods can identify hundreds of putative
protein complexes in a single experiment. The ComplexMap
package provides a comprehensive, end-to-end workflow to process,
analyze, annotate, and visualize such a dataset.
This vignette demonstrates the main analysis workflow using the
high-level createComplexMap() wrapper function. We will use
a dataset of human soluble protein complexes, originally published in
2012 A census of
human soluble protein complexes, to perform a complete analysis from
start to finish. We will then showcase the downstream analysis functions
for interpreting and exploring the results.
Step 1: Loading Data
The ComplexMap package includes the
demoComplexes dataset, a list of 622 putative human protein
complexes. We will also load an example gene set (GMT) file for
functional annotation.
# Load the example complex list shipped with the package
utils::data("demoComplexes", package = "ComplexMap")
# Get the path to the example GMT file and load it
gmtPath <- ComplexMap::getExampleGmt()
biocartaGmt <- ComplexMap::getGmtFromFile(gmtPath, verbose = FALSE)Step 2: Running the Main Workflow
The core of the package is the createComplexMap()
function. This single wrapper function handles all the essential
processing steps:
- Quality Control & Refinement: Filters complexes by size and merges highly redundant ones (defaulting to Jaccard similarity > 0.90).
- Functional Enrichment: Annotates each complex with biological functions, prioritizing specificity.
- Network Construction: Builds a similarity network based on shared proteins (physical) and functions.
- Attribute & Topology Generation: Calculates node colors, sizes, and layout coordinates for visualization.
We can pass parameters to the underlying functions directly into this wrapper.
# Run the entire workflow with a single command
complexMapObject <- ComplexMap::createComplexMap(
complexList = demoComplexes,
gmt = biocartaGmt,
mergeThreshold = 0.90, # Strict merging to preserve variants
alpha = 0.75, # Prioritize physical composition in layout
verbose = TRUE
)
#> --- Starting ComplexMap Workflow ---
#> Parameters: similarity='jaccard', alpha=0.75 (Diversity Priority)
#>
#> Step 1: Refining complex list...
#>
#> --- Refining Input Complex List (Minimal Merging Strategy) ---
#> Filtered 112 complexes by size. Retaining 510.
#> Identifying merge groups: method='jaccard', threshold >= 0.90...
#> Found 0 redundancy groups. Merging 510 complexes into 510.
#>
#> --- Refinement Complete ---
#>
#> Step 2: Running enrichment analysis...
#> Running enrichment for 510 complexes (Cutoff: 0.05)...
#> Annotation complete. Found significant terms for 239 complexes.
#>
#> Step 3: Building complex network...
#> Building complex network (combined mode, alpha=0.75)...
#> Processing 129795 pairs using 7 cores...
#> Network built: 1390 edges retained.
#>
#> Step 4: Generating node attributes...
#> Generating node attributes (prioritizing functional specificity)...
#> -> Clustering 231 terms using co-occurrence (jaccard)
#> Metric: 'jaccard' with unit: 'log'; comparing: 231 vectors
#> -> Generating diverse palette: 25 functional domains (Average Linkage).
#>
#> Step 5: Computing map topology...
#> Computing map topology (layout and centrality)...
#> Topology computation complete.
#>
#> --- ComplexMap Workflow Complete ---The output, complexMapObject, is a formal
ComplexMap S3 object that contains the complete results of
the analysis. Printing the object gives a high-level systems biology
dashboard.
# Print the object to see a summary
complexMapObject
#> # ComplexMap Object (Physical-First Layout)
#> # ── Physical Structure: 510 nodes, 1390 edges (2.73 edges/node)
#> # ── Functional Landscape:
#> # • Diversity: 118 distinct functional domains (colors)
#> # • Coverage: 46.9% of complexes annotated
#> # ── Accessors: `getNodeTable()`, `getEdgeTable()`
#> # ── Analysis: `summarizeThemes()` to identify physical machines.Step 3: Summarizing Biological Themes
A key goal of the map is to identify major biological themes. The
summarizeThemes() function uses community detection
algorithms to find densely connected network modules (physical
neighborhoods) and provides a summary.
# To get the summary table, we set add_to_object = FALSE
themeSummary <- ComplexMap::summarizeThemes(
complexMapObject,
verbose = FALSE,
add_to_object = FALSE
)
# Display the top 10 largest themes
themeSummary %>%
dplyr::arrange(dplyr::desc(nodeCount)) %>%
utils::head(10) %>%
knitr::kable()| themeId | themeLabel | themePurity | nodeCount | edgeCount |
|---|---|---|---|---|
| 1 | BIOCARTA_SALMONELLA_PATHWAY / BIOCARTA_CPSF_PATHWAY | 0.13 | 47 | 121 |
| 5 | BIOCARTA_KREB_PATHWAY / BIOCARTA_ETC_PATHWAY | 0.36 | 38 | 56 |
| 6 | BIOCARTA_CELL2CELL_PATHWAY / BIOCARTA_TEL_PATHWAY | 0.19 | 36 | 71 |
| 9 | BIOCARTA_RANMS_PATHWAY / BIOCARTA_NPC_PATHWAY | 0.29 | 28 | 105 |
| 8 | BIOCARTA_IRES_PATHWAY / BIOCARTA_MCM_PATHWAY | 0.22 | 27 | 76 |
| 14 | BIOCARTA_PROTEASOME_PATHWAY / BIOCARTA_ERAD_PATHWAY | 0.42 | 27 | 95 |
| 4 | BIOCARTA_TID_PATHWAY / BIOCARTA_CARM_ER_PATHWAY | 0.43 | 23 | 52 |
| 16 | BIOCARTA_LIS1_PATHWAY / BIOCARTA_G2_PATHWAY | 0.27 | 23 | 30 |
| 2 | BIOCARTA_CD40_PATHWAY / BIOCARTA_DNAFRAGMENT_PATHWAY | 0.14 | 22 | 45 |
| 7 | BIOCARTA_MTA3_PATHWAY / BIOCARTA_PRC2_PATHWAY | 0.29 | 22 | 39 |
The result is a table listing each theme, its descriptive label (derived from the most frequent functions within the theme), and its size in terms of nodes and edges.
Step 4: Exploring and Querying the Map
The queryMap() function provides a powerful way to
programmatically explore the results.
4.1 Querying for a Specific Protein
Let’s find all complexes that contain the protein “SMAD4”.
# To ensure our example is robust, let's find a protein to query
# that is guaranteed to be in our final, refined map.
nodes <- ComplexMap::getNodeTable(complexMapObject)
if (nrow(nodes) > 0) {
first_protein_list <- nodes$proteins
query_protein <- strsplit(first_protein_list, ",")[[1]][1]
message("Dynamically querying for a protein found in the map: ", query_protein)
protein_complexes <- ComplexMap::queryMap(
complexMapObject,
query = query_protein,
type = "protein"
)
# Show the primary functional domain of the resulting complexes
protein_complexes %>%
dplyr::select(complexId, primaryFunctionalDomain, proteins) %>%
knitr::kable()
}
#> Dynamically querying for a protein found in the map: WDR3| complexId | primaryFunctionalDomain | proteins |
|---|---|---|
| CpxMap_0414 | BIOCARTA_CIRCADIAN_PATHWAY | WDR3,RB1,PNO1,EMG1,CSNK1E,TPTEP2-CSNK1E,DDX49,BYSL |
4.2 Querying for a Specific Complex
We can also retrieve the data for a single complex of interest.
if (nrow(nodes) > 0) {
# Query for the first node in the table
first_complex_id <- nodes$complexId[1]
complex_data <- ComplexMap::queryMap(
complexMapObject,
query = first_complex_id,
type = "complex"
)
dplyr::glimpse(complex_data)
}
#> Rows: 1
#> Columns: 11
#> $ complexId <chr> "CpxMap_0414"
#> $ proteinCount <int> 8
#> $ proteins <chr> "WDR3,RB1,PNO1,EMG1,CSNK1E,TPTEP2-CSNK1E,DDX49…
#> $ primaryFunctionalDomain <chr> "BIOCARTA_CIRCADIAN_PATHWAY"
#> $ topEnrichedFunctions <chr> "BIOCARTA_CIRCADIAN_PATHWAY; BIOCARTA_TERC_PAT…
#> $ colorHex <chr> "#C28434"
#> $ sizeMapping <dbl> 3
#> $ x <dbl> 9.709937
#> $ y <dbl> -0.9679479
#> $ betweenness <dbl> 0.2358685
#> $ degree <dbl> 45Step 5: Visualization
Finally, we can visualize the entire functional landscape.
ComplexMap provides three visualization functions that all
work directly with the ComplexMap object.
First, we extract the final node and edge tables for plotting.
mapLayout <- ComplexMap::getNodeTable(complexMapObject)
networkEdges <- ComplexMap::getEdgeTable(complexMapObject)5.1 Static Plot with a Legend
This version is useful for a clean overview, using a discrete color legend to represent the functional domains.
if (nrow(mapLayout) > 0) {
ComplexMap::visualizeMapWithLegend(mapLayout, networkEdges)
}
#> Visualizing diverse landscape with color legend...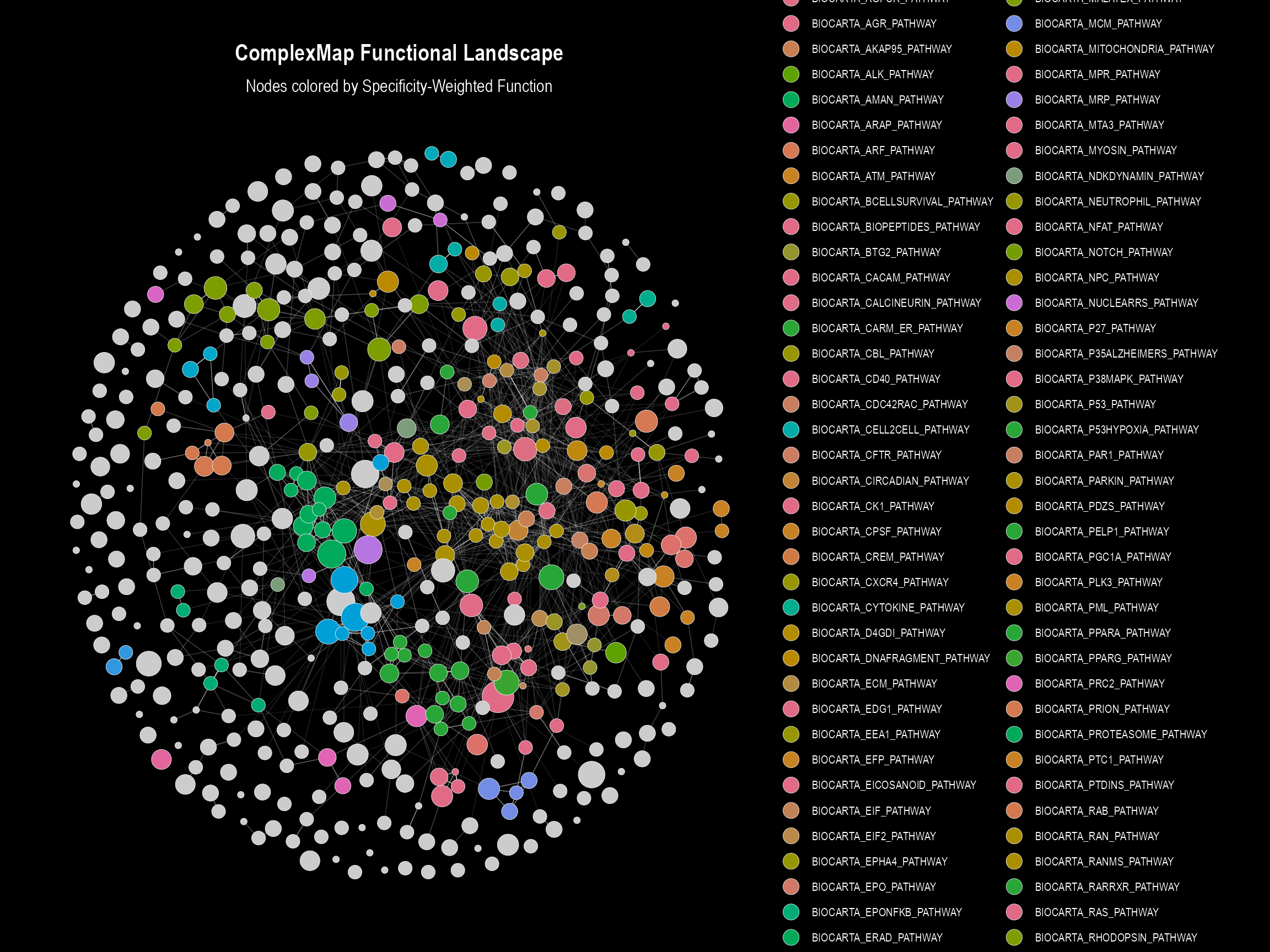
5.2 Interactive Plot
For deep exploration, an interactive HTML widget is ideal. You can zoom, pan, and hover over nodes to see detailed tooltips.
# visNetwork is required for this plot
if (requireNamespace("visNetwork", quietly = TRUE) && nrow(mapLayout) > 0) {
ComplexMap::visualizeMapInteractive(mapLayout, networkEdges)
}
#> Generating interactive visNetwork plot...Conclusion
By using the main createComplexMap() wrapper, a complete
analysis can be run in a single step. The resulting object can then be
easily interpreted, queried, and visualized, providing a user-friendly
and powerful platform for exploring the landscape of protein
complexes.