Qingzhou Zhang
A computational systems biologist
About
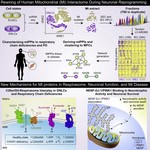
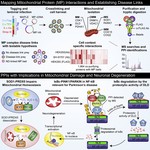
I am a computational systems biologist dedicated to building predictive models of life. I believe that the next era of biological discovery and therapeutic design will be driven by our ability to learn the causal rules that govern complex cellular systems.
My scientific blueprint is to build interpretable AI models from a synthesis of multi-modal data, from single-cell genomics to spatial proteomics. By focusing on perturbation-response experiments, my work moves beyond correlation to infer the causal logic of how cellular networks operate and adapt.
Interests
- Predictive Systems Biology
- Causal Network Inference
- Cancer Therapeutic Resistance
- Tissue Homeostasis & Repair
- Interpretable Machine Learning
Education
-
PhD Biochemistry, 2016-2021
University of Regina, Canada
-
MSc Bioinformatics, 2009-2010
Univeristy of Manchester, England
-
BSc Biotechnology, 2004-2008
Nanjing Agricultural University, P.R. China